A polychaete with a pair of tentacles was found boring the shell of a gastropod in Oshoro Bay, Hokkaido, Japan, about 43°12′N, 140°51′E on 28 May 2012 by Minako Ito and Hikaru Osajima. The polychaete was removed from the shell, photographed alive, and identified by Hiroshi Kajihara as Polydora sp., then fixed in 99% EtOH; the gastropod, identified as Chlorostoma turbinatum Adams, 1853, was also fixed and preserved in 99% EtOH. DNA was extracted from the entire specimen of the polychaete using the silica method, dissolved in 30 µl of deionized water, and has been preserved at –20°C.
PCR amplification of the so called "Folmer region" of the mitochondrial cytochrome c oxidase subunit I (COI) gene, with the primer pair HCO2198 and LCO1490 (Folmer et al. 1994) was unsuccessful.
An about 1000-bp fragment of 28S rDNA was amplified by PCR using LSU5 (5′ -ACCCGCTGAAYTTAAGCA-3′) and LSU3 (5′-TCCTGAGGGAAACTTCGG-3′) (Littlewood 1994). A hot start PCR was performed by a thermal cycler, DNA Engine (Bio-Rad), in a 20-µl reaction volume containing 1-µl of template total DNA (approximately 10–100ng) and 19-µl of premix made with 632-µl deionized water, 80-µl Ex Taq Buffer (TaKara Bio), 64-µl dNTP (each 25 mM), 8-µl each primer (each 10 µM), and 0.1-µl TaKara Ex Taq (5 U/µl, TaKara Bio). Thermal cycling condition comprised an initial denaturation at 95°C for 30 sec; 30 cycles of denaturation at 95°C for 30 sec, annealing at 45°C for 30 sec, and elongation at 72°C for 45°C and a final elongation at 72°C for 7 min.
The PCR product was purified with the silica method (Boom et al. 1990). Both strands were sequenced with a BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) following the manufacturer's protocol, using the above mentioned primer set as the initial PCR amplification, and D2F (5′ -CTTTGAAGAGAGAGTTC-3′) (Littlewood 1994) and 28Z (5′ -CTTGGTCCGTGTTTCAGAC-3′) (Hillis and Dixon 1991) set as internal PCR amplification. Sequencing was performed with ABI Prism 3730 DNA Analyzer (Applied Biosystems). Chromatogram and sequence data were operated with MEGA v5 software (Tamura et al. 2011).
Results
A total of 1045 bp of 28S rDNA sequence was determined from Polydora sp. (see Appendix). A BLAST search (Altschul et al. 1997), as implemented in the NCBI website (http://www.ncbi.nlm.nih.gov/), resulted in that the present sequence showed 92% maximum identity with DQ790059, a 3414-bp strech of partial 28S rDNA sequence from an unidentified Polydora sp. (Struck et al. 2007).
Taxonomy
Phylum Annelida
Class Polychaeta
Order Spionida
Family Spionidae Grube, 1850
Genus Polydora Bosc, 1802
Polydora sp. indet.
(Figs 1–4)
Material examined: ICHU22100158 (non-extant, completely destroyed during DNA extraction).

Fig. 1. Polydora sp., ICHU22100158, photograph of the specimen, partially exposed from the living tube made within the shell of the living gastropod, Chlorostoma turbinatum Adams, 1853.

Fig. 2. Polydora sp., ICHU22100158, removed from the tube, ventral view.
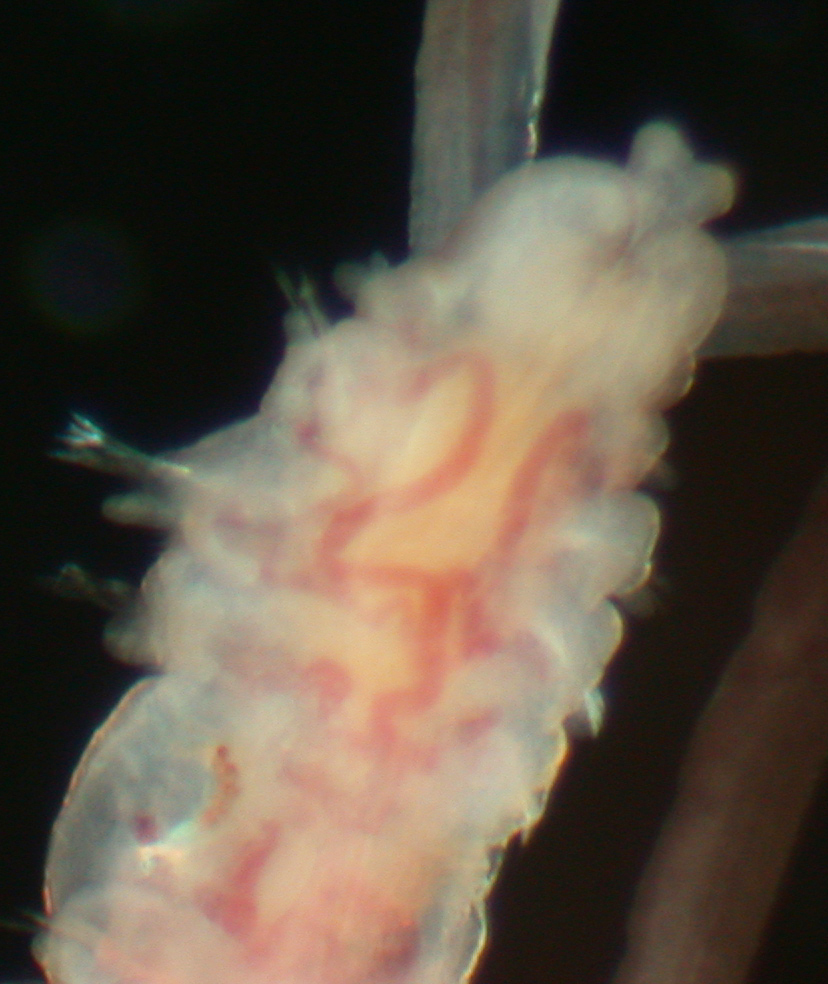
Fig. 3. Polydora sp., ICHU22100158, magnification of head, ventral view.

Fig. 4. Polydora sp., ICHU22100158, magnification of head, dorsal view.
References
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. and Lipman, D. J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402.
Boom, R., Sol, C. J. A., Salimans, M. M. M., Jansen, C. L., Wertheim-van Dillen, P. M. E., and van der Noordaa, J. 1990. Rapid and simple method for purification of nucleic acids. Journal of Clinical Microbiology 28:495–503.
Folmer, O., Black, M., Hoeh, W., Lutz, R. and Vrijenhoek, R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3: 294–299.
Hillis, D. M. and Dixon, M. T. 1991. Ribosomal DNA: molecular evolution and phylogenetic inference. Quarterly Review of Biology 66: 411–453.
Littlewood, D. T. 1994 Molecular phylogenetics of cupped oysters based on partial 28S rRNA gene sequences. Molecular Phylogenetics and Evolution 3: 221–229.
Struck, T. H., Schult, N., Kusen, T., Hickman, E., Bleidorn, C., McHugh, D. and Halanych, K. M. 2007. Annelid phylogeny and the status of Sipuncula and Echiura. BMC Evolutionary Biology 7: 57. doi:10.1186/1471-2148-7-57
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution 28: 2731–2739.
Appendix28S rRNA sequence from ICHU22100158, identified as Polydora sp.
AGGATTCCCTTAGTAACGGCGAGTGAAACGGGAAGAGCCCAGCACCGAATCCCCTGCCTGTGCAGCGGGAACTGTGGTGTTTAGGAGAGCCTGTGGAGCGGCCGGAGGCGTCCGAGTCCTTATGAAAGGGGCTACAGCCAGAGCGGGTGTCAGTCCTCTAGGACGCCACCGTGCCGTTGCTCAGTACCTCCTCGGAGTCGGGTTGTTTGTGAATGCAGCCCAAAGTGGGTGGTAAACTCCATCTAAGGCTAAATACTGGCACGAGACCGATAGCAAACAAGTACCGTGAGGGAAAGTTGAAAAGAACTTTGAAGAGAGAGTTCAAGAGTACGTGAAACCGTATAGAAGTAAACGGATGTGACCGCGTAATGGACCGGCGGGATTCAGTTTTCACGGGTGCGAGAGGCGGCGGGACGGAGACCTTCACCGGCTCCCCTCGTCGTCTGCCCGCTCCGTGGGAATGCACTTTCCGCCGGGACAGCGCCACGACCGGTTTTTCGGCCGCCTAGGCCCGTGGGGAAGGTAGCTCTGCCTCGGCCGAGTATTATAGCCCCGCGGTGTTCCGGCCGGTTGAGAGACCGAGGAATCCTTACCCGCCGCTCGGGGAGACCTGGCGCGTCGGTGGCAACTCTACTGGTGAGGACTGCTCGCAGTGCTCGTCGACCGCTGCCGCCGTTCGTGTCGGGCCCCCTCCTCGAAGCTCAGGGTCAGTGGCGAAAGTTGGTTCCATATCCGACCCGTCTTGAAACACGGACCAAGGAGTCTAACATGTGCGCAAGTCATTGGGTGTTACGAAACCTAAAGGCGCAATGAAAGTAAAGGCTCTCTTACGAGGGCTGAGGTGTGATCCGGTCTCGGCTGGCGCAACACCGGCCCGTCTCGAGGGCTTTGTCCCTGAGGCGGCGCAAGAGCGTACACGTTAGGACCCGAAAGATGGTGACCTATGCCTGAGTAGGTCGAAGCCAGAGGAAACTCTGGTGGAGGACCGTAGCGATTCTGACGTGCAAATCGATCGTCGAACTTGGGTATAGGGGCGAAAGACTAAT