A nudibranch was collected on a stone in slight shore reef in Oshoro Bay, Hokkaido, Japan, about 43°12′N, 140°51′E, on 29 June 2015 by Hanon Yamagchi. The specimen was photographed with a D5200 digital camera (Nikon) by Shinri Tomioka and fixed in 99% EtOH; it was later identified by Hiroshi Kajihara as Flabellina athadona (Eschscholtz, 1831) by the reference to Hamatani (1992) based on the photographs taken in life. Total DNA was extracted from the posterior portion of the body using the silica method (Boom et al. 1990) with some modifications. Extracted DNA was dissolved in 30 µl of deionized water and has been preserved at –20°C. Remaining morphological voucher specimen has been deposited at the Hokkaido University Museum under the catalogue number ICHU2130729 (contact: Dr. Hiroshi Kajihara, kazi@mail.sci.hokudai.ac.jp).
PCR amplification was attempted with the primer pairs LoboF1 (5′-KBTCHACAAAYCAYAARGAYATHGG-3′) and LoboR1 (5′-TGTTTYTTYGGWCAYCCWGARGTTTA-3′) (Lobo et al. 2013) for the mitochondrial cytochrome c oxidase subunit I gene (COI) and 16S ar-L (5′-CGCCTGTTTATCAAAAACAT-3′) and br-H (5′-CCGGTCTGAACTCAGATCACGT-3′) (Palumbi et al. 1991) for the 16S rRNA gene . PCR products were visualized by electrophoresis in 1% agarose gel. Of the two gene markers that were attempted, only 16S rRNA gene was confirmed to be successfully amplified. The PCR was performed by a thermal cycler, 2720 Thermal Cycler (Applied Biosystems), in a 20-µl reaction volume containing 1 µl of template total DNA (approximately 10–100 ng) and 19 µl of premix made with 632-µl deionized water, 80-µl Ex Taq Buffer (TaKara Bio), 64-µl dNTP (each 25 mM), 8-µl each primer (each 10 µM), and 0.1-µl TaKara Ex Taq (5 U/µl,TaKara Bio). Thermal cycling condition comprised an initial denaturation at 95°C for 30 sec; 30 cycles of denaturation at 95°C for 30 sec, annealing at 45°C for 30 sec, and elongation at 72°C for 45°C and a final elongation at 72°C for 7 min.
The PCR products of the 16S rRNA gene was purified with the silica method (Boom et al. 1990). Both strands were sequenced with a BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) following the manufacturer's protocol, using the same primer set as the initial PCR amplification. Sequencing was performed with ABI Prism 3730 DNA Analyzer (Applied Biosystems). Chromatogram and sequence data were operated with MEGA v5.2 software (Tamura et al. 2011).
Results
A total of 423 bp of the mitochondrial 16S rRNA gene sequence was determined from ICHU2130729, identified as Hermissenda crassicornis (see Appendix). A nucleotide BLAST search (Altschul et al. 1997) at the NCBI website (https://blast.ncbi.nlm.nih.gov/) resulted in that our sequence from Oshoro showed 99% similarity with JQ699554 (E value = 0.0; 97% query covorage), a sequence from a specimen identified as the same species from the Pacific coast of the USA (Churchill et al. 2013).
Taxonomy
Phylum Mollusca
Class Gastropoda
Order Nudibranchia
Family Facelinidae
Genus Hermissenda Bergh, 1879
Hermissenda crassicornis (Eschscholtz, 1831)
[Japanese name: emura-mino-umiushi]
(Fig. 1)
Hermissenda crassicornis: Hamatani (1992), p. 289, pl. 58-6.
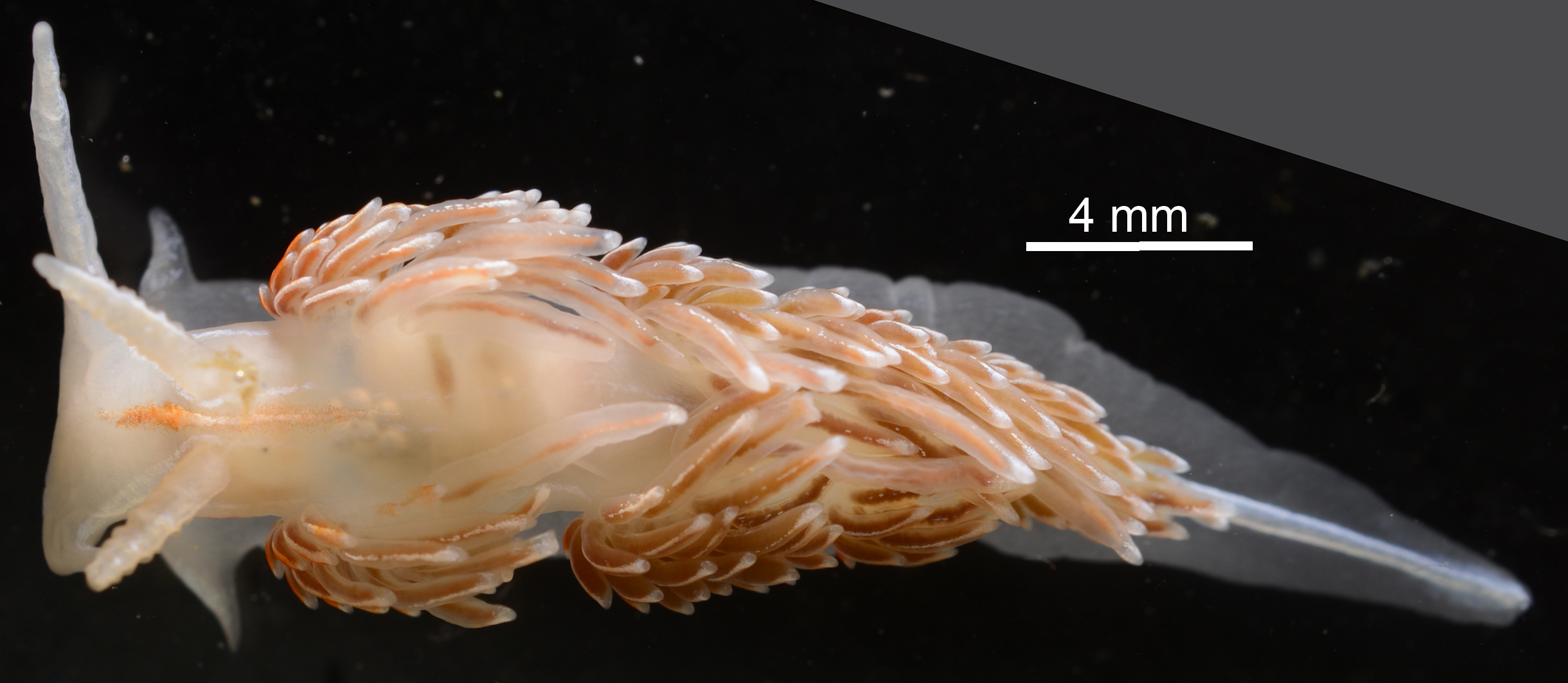
Fig. 1. Hermissenda crassicornis (Eschscholtz, 1831) (ICHU2130729) from Oshoro Bay, photographed by Shinri Tomioka.
References
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402.
Boom, R., Sol, C. J., Salimans, M. M., Jansen, C. L., Wertheim-van Dillen, P. M., and Van der Noordaa, J. 1990. Rapid and simple method for purification of nucleic acids. Journal of Clinical Microbiology 28: 495–503.
Churchill, C. K. C., Alejandrino, A., Valdés, Á., and Foighil, D. Ó. 2013. Parallel changes in genital morphology delineate cryptic diversification of planktonic nudibranchs. Proceedings of the Royal Society B 280: 20131224. http://dx.doi.org/10.1098/rspb.2013.1224
Hamatani, I. 1992. Opisthobranchia. Pp. 267–290. In: Nishimura, S. (Ed.) Guide to Seashore Animals of Japan with Clor Pictures and Keys, Vol. I. Hoikusha, Osaka. [In Japanese]
Lobo, J., Costa, P. M., Teixeira, M. A. L., Ferreira, M. S. G., Costa, M. H, and Costa, F. O. 2013. Enhanced primers for amplification of DNA barcodes from a broad range of marine metazoans. BMC Ecology 13:34. DOI: 10.1186/1472-6785-13-34.
Palumbi, S., Martin, A., Romano, S., McMillan, W. O., Stice, L., Grabowski, G. 1991. The Simple Fools Guide to PCR, Ver. 2. Department of Zoology and Kewalo Marine Laboratory, University of Hawaii, Honolulu, 45 pp.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. 2011. MEGA5: molecullar evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739.
AppendixA 423-bp partial sequence of the mitochondrial 16S rRNA gene determined from ICHU2130729, identified as Hermissenda crassicornis.
AGCCACAGGCTTATATTTGTGGTTCAACCTGCCCTATGAATTTTAATGGCCGCAGTACTCTGACTGTGCTAAGGTAGCGTAATCAAGTTGGCTTTTAAATGGAGTCGGGAATGAATGGGAAAACCCCGGTTTAGCTGTCGAGGCTTACTTACTTGAATTTGCTTTTTAAGTGAAAAAGCTTAAAAGAAAAAAAAAGACGAGAAGACCCTTAGAACTTTAATTATAGAACATCTAACGTTTTTATTGGGGCGATAAAGAAACATTGAAACTTTCTGTAAAAACAAGCCGATTTTATTATGGGAGAATAAGTTACCTGAGGGATAACAGCATAATTTTATAATTAAGTTTGTGACCTCGATGTTGGACTAGGAAACTAATAGGCTAGCCGTTTATTATGTGAGTTCTGTTCGAACTATTATTCCT